from that_ml_library.data_preprocess import *
from that_ml_library.chart_plotting import *
from that_ml_library.ml_helpers import *End-to-end tutorial - Classification
In this tutorial, we will use the Pima Indian dataset https://www.kaggle.com/datasets/uciml/pima-indians-diabetes-database. The objective of the dataset is to diagnostically predict whether or not a patient has diabetes, based on certain diagnostic measurements included in the dataset.
Load data
import pandas as pd
import numpy as npdf = pd.read_csv('https://raw.githubusercontent.com/anhquan0412/dataset/main/diabetes.csv')
print(df.shape)
display(df.sample(7))(768, 9)| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 92 | 7 | 81 | 78 | 40 | 48 | 46.7 | 0.261 | 42 | 0 |
| 54 | 7 | 150 | 66 | 42 | 342 | 34.7 | 0.718 | 42 | 0 |
| 288 | 4 | 96 | 56 | 17 | 49 | 20.8 | 0.340 | 26 | 0 |
| 90 | 1 | 80 | 55 | 0 | 0 | 19.1 | 0.258 | 21 | 0 |
| 98 | 6 | 93 | 50 | 30 | 64 | 28.7 | 0.356 | 23 | 0 |
| 397 | 0 | 131 | 66 | 40 | 0 | 34.3 | 0.196 | 22 | 1 |
| 574 | 1 | 143 | 86 | 30 | 330 | 30.1 | 0.892 | 23 | 0 |
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KBTrain test split and preprocessing
from sklearn.model_selection import train_test_splitX_train, X_test, y_train, y_test = train_test_split(df.iloc[:,:-1], df.Outcome,
test_size=0.2,
random_state=42,
stratify=df.Outcome)X_train.describe()| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| count | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 |
| mean | 3.819218 | 120.908795 | 69.442997 | 20.776873 | 78.666124 | 31.973290 | 0.477428 | 33.366450 |
| std | 3.314148 | 31.561093 | 18.402581 | 15.856433 | 107.736572 | 7.861364 | 0.330300 | 11.833438 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.084000 | 21.000000 |
| 25% | 1.000000 | 99.000000 | 62.500000 | 0.000000 | 0.000000 | 27.500000 | 0.245000 | 24.000000 |
| 50% | 3.000000 | 117.000000 | 72.000000 | 23.000000 | 40.500000 | 32.300000 | 0.382500 | 29.000000 |
| 75% | 6.000000 | 140.000000 | 80.000000 | 32.000000 | 130.000000 | 36.500000 | 0.639250 | 41.000000 |
| max | 17.000000 | 199.000000 | 122.000000 | 99.000000 | 744.000000 | 67.100000 | 2.329000 | 81.000000 |
For this dataset, we only need to perform scaling on numerical features, as there’s no categorical features to process. Note that scaling of numerical data is required for linear model such as linear regression, and not required for tree model, though it doesn’t hurt the performance of such tree model. For simplicity, we will scale the numerical features for all the models we are going to run below.
num_cols = df.columns[:-1].tolist()
print(num_cols)['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age']For Blood Pressure and Skin Thickness, a value of 0 does not make sense, so we will consider these as missing values. The strategy for filling in the missing values will be ‘median’
miss_cols = ['BloodPressure','SkinThickness']X_train_processed,X_test_processed = preprocessing_general(X_train,X_test,
miss_cols=miss_cols,
missing_vals=0,strategies='median',
num_cols=num_cols,
scale_methods='minmax')X_train_processed.describe()| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| count | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 | 614.000000 |
| mean | 0.224660 | 0.607582 | 0.569205 | 0.209867 | 0.105734 | 0.476502 | 0.175246 | 0.206107 |
| std | 0.194950 | 0.158598 | 0.150841 | 0.160166 | 0.144807 | 0.117159 | 0.147127 | 0.197224 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.058824 | 0.497487 | 0.512295 | 0.000000 | 0.000000 | 0.409836 | 0.071715 | 0.050000 |
| 50% | 0.176471 | 0.587940 | 0.590164 | 0.232323 | 0.054435 | 0.481371 | 0.132962 | 0.133333 |
| 75% | 0.352941 | 0.703518 | 0.655738 | 0.323232 | 0.174731 | 0.543964 | 0.247327 | 0.333333 |
| max | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
Machine Learning Model Experiments
from sklearn.model_selection import StratifiedKFoldcv = StratifiedKFold(5)Sklearn Logistic Regression
df_train = X_train_processed.copy()
df_train['has_diabetes'] = y_trainget_vif(df_train,plot_corr=True)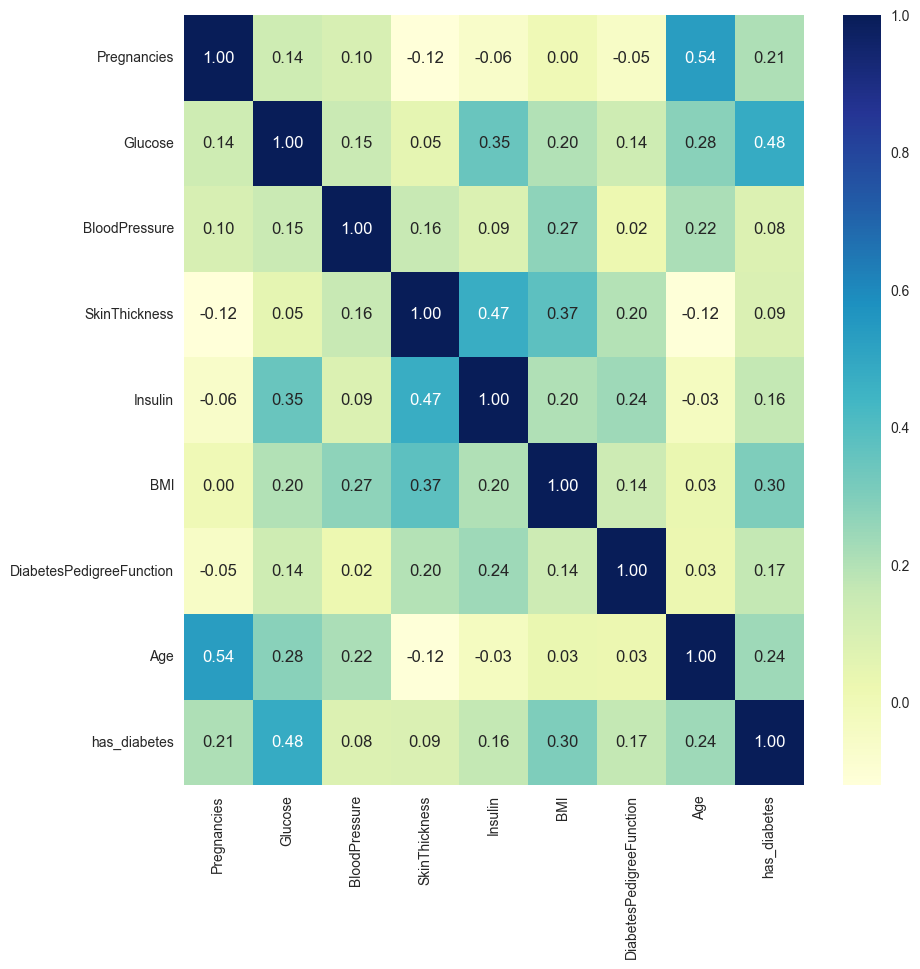
const 40.937975
Pregnancies 1.441405
Glucose 1.573457
BloodPressure 1.152426
SkinThickness 1.529585
Insulin 1.538192
BMI 1.343058
DiabetesPedigreeFunction 1.097619
Age 1.577323
has_diabetes 1.451467
dtype: float64With VIF, we can see that there’s not much colinearity in this dataset. Note that VIF >5 or >10 means high colinearity
run_logistic_regression(X_train_processed,y_train,
multi_class='multinomial',
solver='newton-cg',max_iter=10000)| Features | Coefficients | Coefficients Exp | |
|---|---|---|---|
| 0 | Intercept | -4.233888 | 0.014496 |
| 1 | Pregnancies | 0.976679 | 2.655623 |
| 2 | Glucose | 3.679170 | 39.613487 |
| 3 | BloodPressure | -0.679500 | 0.506870 |
| 4 | SkinThickness | 0.217158 | 1.242540 |
| 5 | Insulin | -0.473694 | 0.622698 |
| 6 | BMI | 3.106698 | 22.347132 |
| 7 | DiabetesPedigreeFunction | 0.881984 | 2.415688 |
| 8 | Age | 0.461467 | 1.586400 |
----------------------------------------------------------------------------------------------------
Log loss: 0.46620685663024525
----------------------------------------------------------------------------------------------------
precision recall f1-score support
0 0.81 0.90 0.85 400
1 0.76 0.60 0.67 214
accuracy 0.79 614
macro avg 0.78 0.75 0.76 614
weighted avg 0.79 0.79 0.79 614
LogisticRegression(max_iter=10000, multi_class='multinomial', penalty=None,
random_state=0, solver='newton-cg')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression(max_iter=10000, multi_class='multinomial', penalty=None,
random_state=0, solver='newton-cg')Interpretation of coefficients in logistic regression: For a coefficient β1, given a one unit increase in one of the variables (say X1), the odds of X (p(X)/1-p(X)) will be multiplied by e^β1.
In this case, we can tell from coefficient of Glucose that: the odd of having diabetes will be multiplied by ~40 if Glucose level is increased by 1
Statsmodel
run_multinomial_statmodel(X_train_processed,y_train,add_constant=True)Optimization terminated successfully.
Current function value: 0.466207
Iterations 6
MNLogit Regression Results
==============================================================================
Dep. Variable: Outcome No. Observations: 614
Model: MNLogit Df Residuals: 605
Method: MLE Df Model: 8
Date: Mon, 19 Feb 2024 Pseudo R-squ.: 0.2789
Time: 11:43:30 Log-Likelihood: -286.25
converged: True LL-Null: -396.97
Covariance Type: nonrobust LLR p-value: 1.909e-43
============================================================================================
Outcome=1 coef std err z P>|z| [0.025 0.975]
--------------------------------------------------------------------------------------------
const -8.4678 0.805 -10.523 0.000 -10.045 -6.891
Pregnancies 1.9534 0.624 3.129 0.002 0.730 3.177
Glucose 7.3584 0.855 8.609 0.000 5.683 9.034
BloodPressure -1.3590 0.733 -1.855 0.064 -2.795 0.077
SkinThickness 0.4343 0.788 0.551 0.581 -1.110 1.978
Insulin -0.9474 0.856 -1.107 0.268 -2.625 0.730
BMI 6.2135 1.150 5.405 0.000 3.960 8.467
DiabetesPedigreeFunction 1.7640 0.755 2.335 0.020 0.283 3.245
Age 0.9229 0.616 1.499 0.134 -0.284 2.130
============================================================================================
----------------------------------------------------------------------------------------------------
Log loss: 0.46620685662395556
----------------------------------------------------------------------------------------------------
precision recall f1-score support
0 0.81 0.90 0.85 400
1 0.76 0.60 0.67 214
accuracy 0.79 614
macro avg 0.78 0.75 0.76 614
weighted avg 0.79 0.79 0.79 614
<statsmodels.discrete.discrete_model.MNLogit>Decision Tree
Validation Curve
from sklearn.tree import DecisionTreeClassifierdt = DecisionTreeClassifier(criterion='entropy',random_state=42,class_weight=None)
cv = StratifiedKFold(5)
plot_validation_curve(dt,'Val Curve - Decision Tree - PID',X_train_processed,y_train,
cv=cv,param_range=np.arange(1,20,1),param_name='max_depth',scoring='f1_macro',
n_jobs=-1,figsize=(6,4))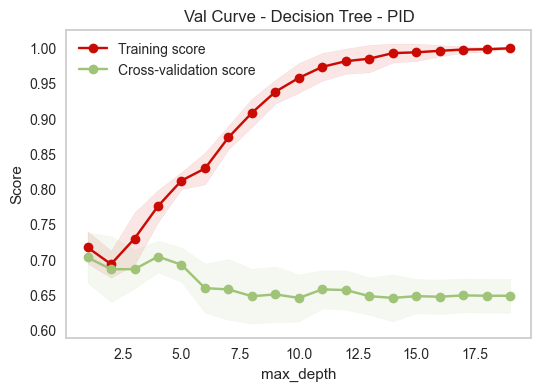
This is the validation curve for hyperparameter max depth, which can be used to prune a decision tree.
For max depth, we can observe the overfitting behavior when max depth is too high, which means the tree is able to make unlimited splits to the extreme of classifying each of the training data points correctly, thus failing to generalize. On the other hand, a small depth is enough to achieve a good CV score (from depth 1 to 5). This is a telling sign that some features have strong signals to predict diabetes than others.
Learning curve
np.linspace(0.1,1,20)array([0.1 , 0.14736842, 0.19473684, 0.24210526, 0.28947368,
0.33684211, 0.38421053, 0.43157895, 0.47894737, 0.52631579,
0.57368421, 0.62105263, 0.66842105, 0.71578947, 0.76315789,
0.81052632, 0.85789474, 0.90526316, 0.95263158, 1. ])dt = DecisionTreeClassifier(criterion='entropy',random_state=42,class_weight=None,min_samples_leaf=27)
plot_learning_curve(dt,'DT Learning Curve',X_train_processed,y_train,cv=cv,scoring='f1_macro',
train_sizes=np.linspace(0.1,1,20))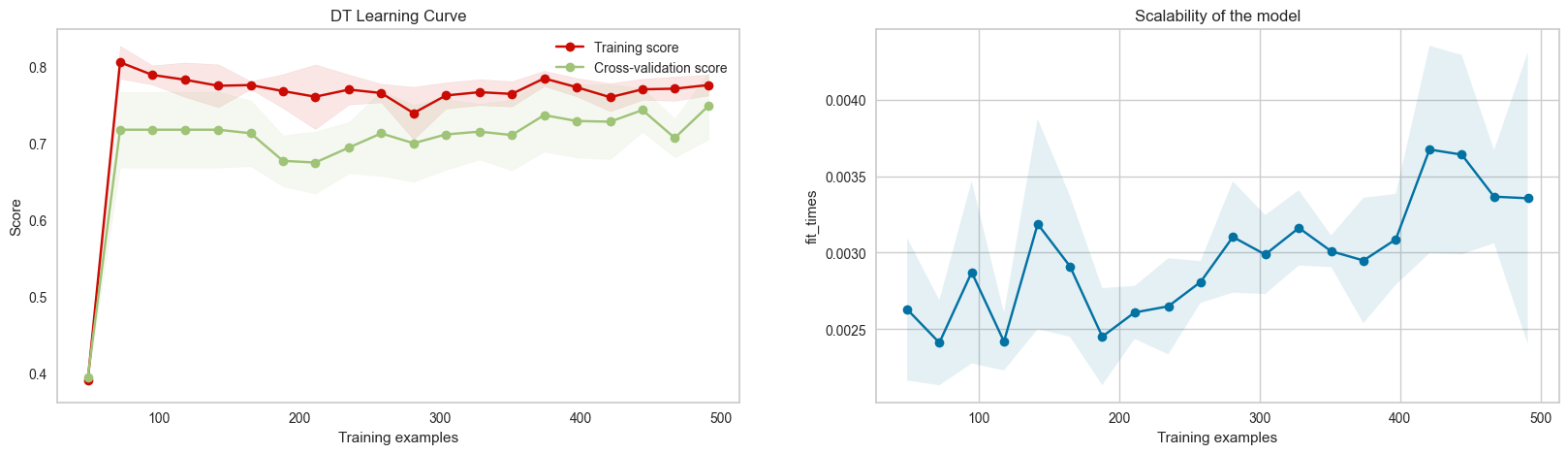
Based on the learning curve for the best tuned decision tree, more data is better for the model to learn, as training score seems to convert, and CV score still slowly increases. However, as the gap between CV score and train score is closing, adding lots of data might not be beneficial. For the scalability of the model, the relationship between training time and fit time is linear, which makes sense as more data allows tree to grow more, which takes more time to fit and predict.
Tree plotting
Using Dtreeviz
dt = DecisionTreeClassifier(criterion='entropy',random_state=42,class_weight=None,max_depth=3)
dt.fit(X_train_processed,y_train)
plot_tree_dtreeviz(dt,X_train_processed,y_train,target_name='has_diabetes',class_names=['no','yes'],
fancy=True,scale=1.0)/home/quan/anaconda3/envs/dev/lib/python3.10/site-packages/sklearn/base.py:439: UserWarning: X does not have valid feature names, but DecisionTreeClassifier was fitted with feature namesGlucose alone can make CV F1 score to reach 0.704, which is not far from the CV score of the best tuned model. With domain knowledge, we can confirm that high glucose is the number one indicator for diabetes, because diabetes patients are unable to effectively use insulin to break down glucose.
Using Sklearn tree plot
dt = DecisionTreeClassifier(criterion='entropy',random_state=42,class_weight=None,max_depth=3)
dt.fit(X_train_processed,y_train)
plot_classification_tree_sklearn(dt,feature_names=X_train_processed.columns.values,
class_names=['no','yes'],
rotate=True,fname='tree_depth_3_PID')Type 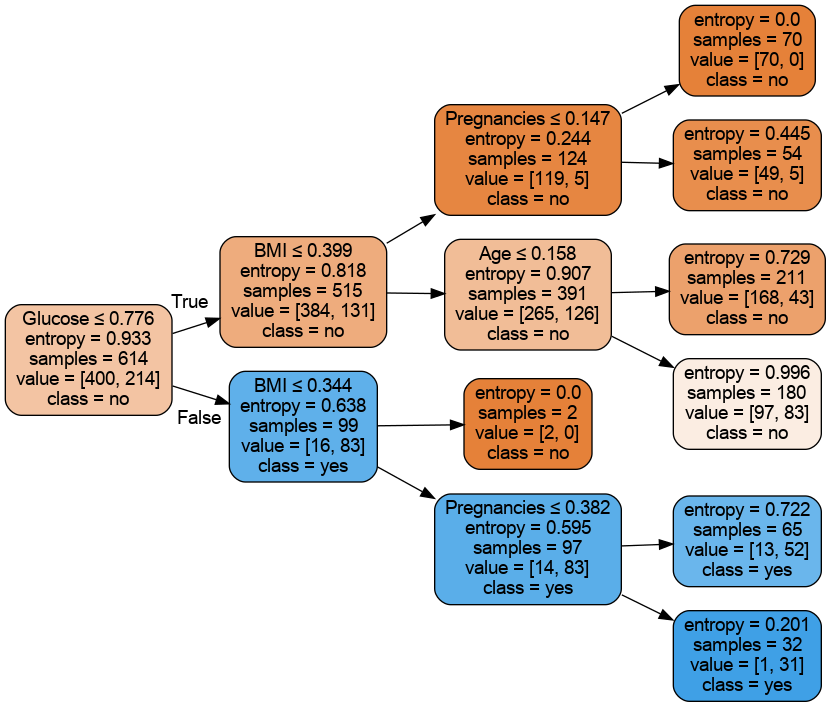

Model finetuning process
from sklearn.ensemble import RandomForestClassifierparam_grid = {
'n_estimators': np.arange(2,80),
'min_samples_leaf': np.arange(1,100),
'max_features': [0.3,0.4,0.5,0.6,0.7,0.8,0.9,1],
}
search_cv = tune_sklearn_model('RandomForest',param_grid,X_train_processed,y_train,
is_regression=False,
custom_cv=5,
scoring='f1_macro',
random_cv_iter=100,
rank_show=7,
show_split_scores=True)Fitting 5 folds for each of 100 candidates, totalling 500 fits
----------
Rank 1
Params: {'n_estimators': 4, 'min_samples_leaf': 23, 'max_features': 0.7}
Train scores: [0.77, 0.76, 0.78, 0.76, 0.78]
Mean train score: 0.768 +- 0.008
Test scores: [0.73, 0.77, 0.71, 0.7, 0.77]
Mean test score: 0.738 +- 0.029
----------
Rank 2
Params: {'n_estimators': 41, 'min_samples_leaf': 5, 'max_features': 0.5}
Train scores: [0.88, 0.87, 0.86, 0.88, 0.87]
Mean train score: 0.872 +- 0.009
Test scores: [0.71, 0.78, 0.71, 0.71, 0.77]
Mean test score: 0.737 +- 0.031
----------
Rank 3
Params: {'n_estimators': 31, 'min_samples_leaf': 10, 'max_features': 0.6}
Train scores: [0.83, 0.81, 0.82, 0.82, 0.81]
Mean train score: 0.819 +- 0.011
Test scores: [0.72, 0.77, 0.7, 0.69, 0.79]
Mean test score: 0.737 +- 0.040
----------
Rank 4
Params: {'n_estimators': 20, 'min_samples_leaf': 28, 'max_features': 0.5}
Train scores: [0.77, 0.76, 0.78, 0.76, 0.78]
Mean train score: 0.770 +- 0.008
Test scores: [0.75, 0.76, 0.71, 0.68, 0.78]
Mean test score: 0.735 +- 0.037
----------
Rank 5
Params: {'n_estimators': 18, 'min_samples_leaf': 94, 'max_features': 0.7}
Train scores: [0.73, 0.73, 0.74, 0.75, 0.72]
Mean train score: 0.735 +- 0.011
Test scores: [0.74, 0.76, 0.7, 0.71, 0.77]
Mean test score: 0.735 +- 0.026
----------
Rank 6
Params: {'n_estimators': 35, 'min_samples_leaf': 61, 'max_features': 0.8}
Train scores: [0.74, 0.74, 0.77, 0.75, 0.73]
Mean train score: 0.746 +- 0.011
Test scores: [0.8, 0.76, 0.69, 0.67, 0.76]
Mean test score: 0.734 +- 0.045
----------
Rank 7
Params: {'n_estimators': 35, 'min_samples_leaf': 8, 'max_features': 0.5}
Train scores: [0.84, 0.82, 0.83, 0.84, 0.82]
Mean train score: 0.832 +- 0.008
Test scores: [0.75, 0.76, 0.7, 0.71, 0.76]
Mean test score: 0.734 +- 0.027
----------
Default Params
Mean train score: 1.0 +- 0.0
Mean test score: 0.731 +- 0.04[Parallel(n_jobs=-1)]: Using backend LokyBackend with 24 concurrent workers.
[Parallel(n_jobs=-1)]: Done 5 out of 5 | elapsed: 0.2s finishedparams_3D_heatmap(search_cv,'n_estimators','min_samples_leaf','max_features','f1_macro')By looking at the 3D map of these three hyperparameters, we might need to exclude max_features of 1 and min_samples_leaf that is more than 80. In fact, max_features of 0.8 and 0.9 shows the most promising set of hyperparameters, thus we can now simplied our search space to this:
param_grid = {
'n_estimators': np.arange(2,80),
'min_samples_leaf': np.arange(1,85),
'max_features': [0.8,0.9],
}
search_cv = tune_sklearn_model('RandomForest',param_grid,X_train_processed,y_train,custom_cv=5,
scoring='f1_macro',
random_cv_iter=200,
rank_show=2,
show_split_scores=True)Fitting 5 folds for each of 200 candidates, totalling 1000 fits
----------
Rank 1
Params: {'n_estimators': 12, 'min_samples_leaf': 12, 'max_features': 0.8}
Train scores: [0.8, 0.8, 0.81, 0.82, 0.8]
Mean train score: 0.806 +- 0.010
Test scores: [0.74, 0.79, 0.7, 0.7, 0.79]
Mean test score: 0.743 +- 0.041
----------
Rank 2
Params: {'n_estimators': 33, 'min_samples_leaf': 23, 'max_features': 0.8}
Train scores: [0.78, 0.76, 0.77, 0.77, 0.76]
Mean train score: 0.769 +- 0.007
Test scores: [0.75, 0.77, 0.71, 0.69, 0.79]
Mean test score: 0.742 +- 0.038
----------
Default Params
Mean train score: 1.0 +- 0.0
Mean test score: 0.731 +- 0.04[Parallel(n_jobs=-1)]: Using backend LokyBackend with 24 concurrent workers.
[Parallel(n_jobs=-1)]: Done 5 out of 5 | elapsed: 0.2s finishedparams_2D_heatmap(search_cv,'n_estimators','min_samples_leaf','f1_macro',
figsize=(20,10),min_hm=0.7,max_hm=None)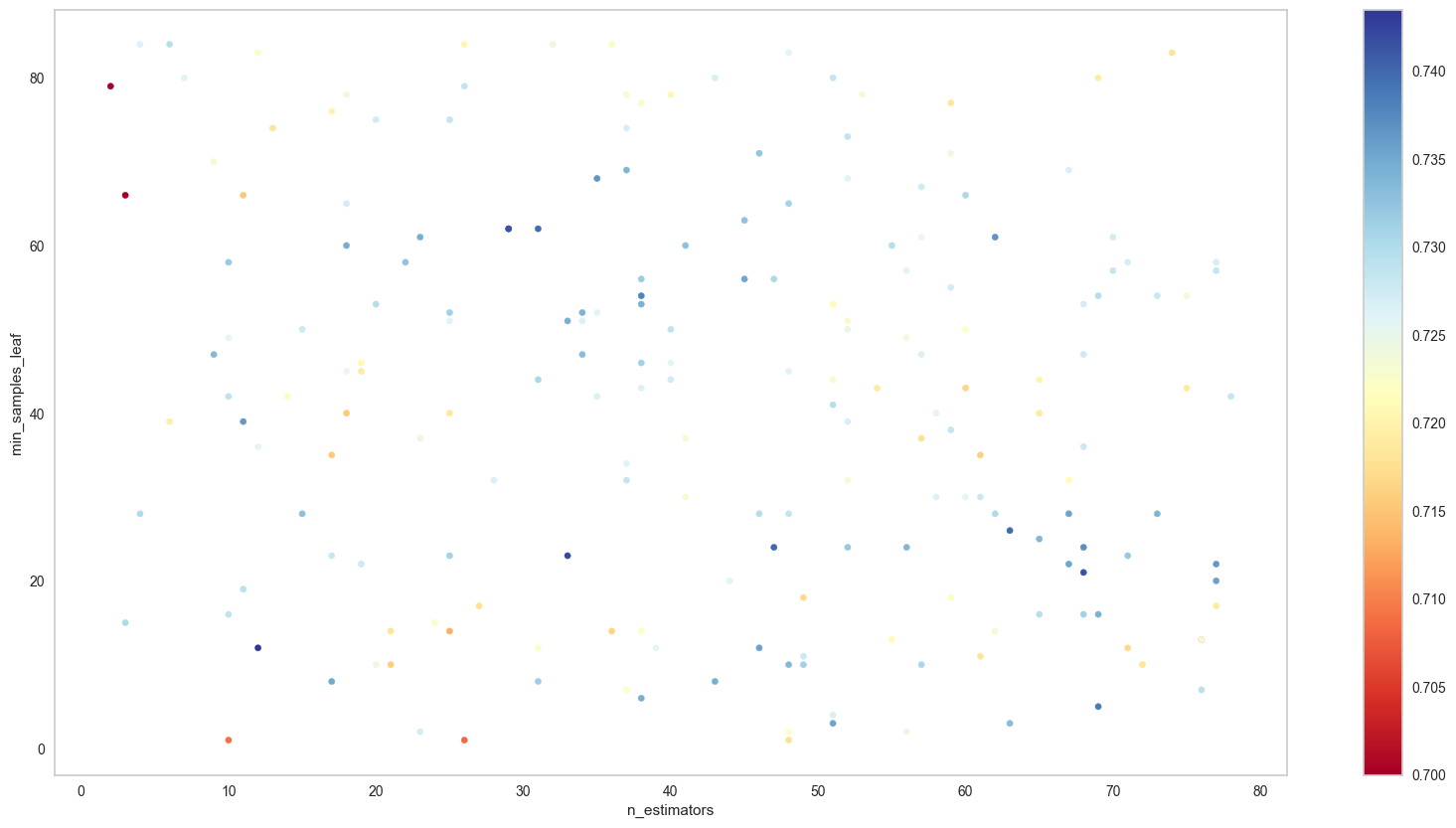
We will now focus on the area near the bottom right of the graph, for grid search
param_grid = {
'n_estimators': np.arange(62,73),
'min_samples_leaf': np.arange(20,26),
'max_features': [0.9],
}
search_cv = tune_sklearn_model('RandomForest',
param_grid,
X_train_processed,
y_train,
scoring='f1_macro',
custom_cv=5,
rank_show=5)Fitting 5 folds for each of 66 candidates, totalling 330 fits
----------
Rank 1
Params: {'max_features': 0.9, 'min_samples_leaf': 25, 'n_estimators': 72}
Train scores: [0.77, 0.75, 0.78, 0.76, 0.77]
Mean train score: 0.767 +- 0.009
Test scores: [0.74, 0.78, 0.7, 0.72, 0.78]
Mean test score: 0.745 +- 0.033
----------
Rank 1
Params: {'max_features': 0.9, 'min_samples_leaf': 25, 'n_estimators': 63}
Train scores: [0.78, 0.75, 0.77, 0.77, 0.77]
Mean train score: 0.765 +- 0.009
Test scores: [0.74, 0.78, 0.7, 0.72, 0.78]
Mean test score: 0.745 +- 0.033
----------
Rank 1
Params: {'max_features': 0.9, 'min_samples_leaf': 25, 'n_estimators': 65}
Train scores: [0.78, 0.75, 0.78, 0.76, 0.77]
Mean train score: 0.766 +- 0.009
Test scores: [0.74, 0.78, 0.7, 0.72, 0.78]
Mean test score: 0.745 +- 0.033
----------
Rank 1
Params: {'max_features': 0.9, 'min_samples_leaf': 25, 'n_estimators': 62}
Train scores: [0.77, 0.75, 0.77, 0.77, 0.77]
Mean train score: 0.765 +- 0.008
Test scores: [0.74, 0.78, 0.7, 0.72, 0.78]
Mean test score: 0.745 +- 0.033
----------
Rank 5
Params: {'max_features': 0.9, 'min_samples_leaf': 25, 'n_estimators': 67}
Train scores: [0.78, 0.75, 0.77, 0.76, 0.77]
Mean train score: 0.766 +- 0.011
Test scores: [0.74, 0.78, 0.7, 0.71, 0.78]
Mean test score: 0.743 +- 0.034
----------
Default Params
Mean train score: 1.0 +- 0.0
Mean test score: 0.731 +- 0.04[Parallel(n_jobs=-1)]: Using backend LokyBackend with 24 concurrent workers.
[Parallel(n_jobs=-1)]: Done 5 out of 5 | elapsed: 0.2s finishedFeature importance (permutation technique)
params= {'max_features': 0.9, 'min_samples_leaf': 25, 'n_estimators': 62}
# Mean train score: 0.765 +- 0.008
# Mean test score: 0.745 +- 0.033# # turn off warnings
# def warn(*args, **kwargs):
# pass
# import warnings
# warnings.warn = warnfinal_model= run_sklearn_model('RandomForest',
params,
X_train_processed,y_train,
is_regression=False,
class_names=['no','yes'],
test_split=None,
seed=42,
plot_fea_imp=True)------------------------------ Train set ------------------------------
Log loss: 0.4134397147185217
precision recall f1-score support
no 0.82 0.90 0.86 400
yes 0.78 0.63 0.70 214
accuracy 0.81 614
macro avg 0.80 0.77 0.78 614
weighted avg 0.80 0.81 0.80 614
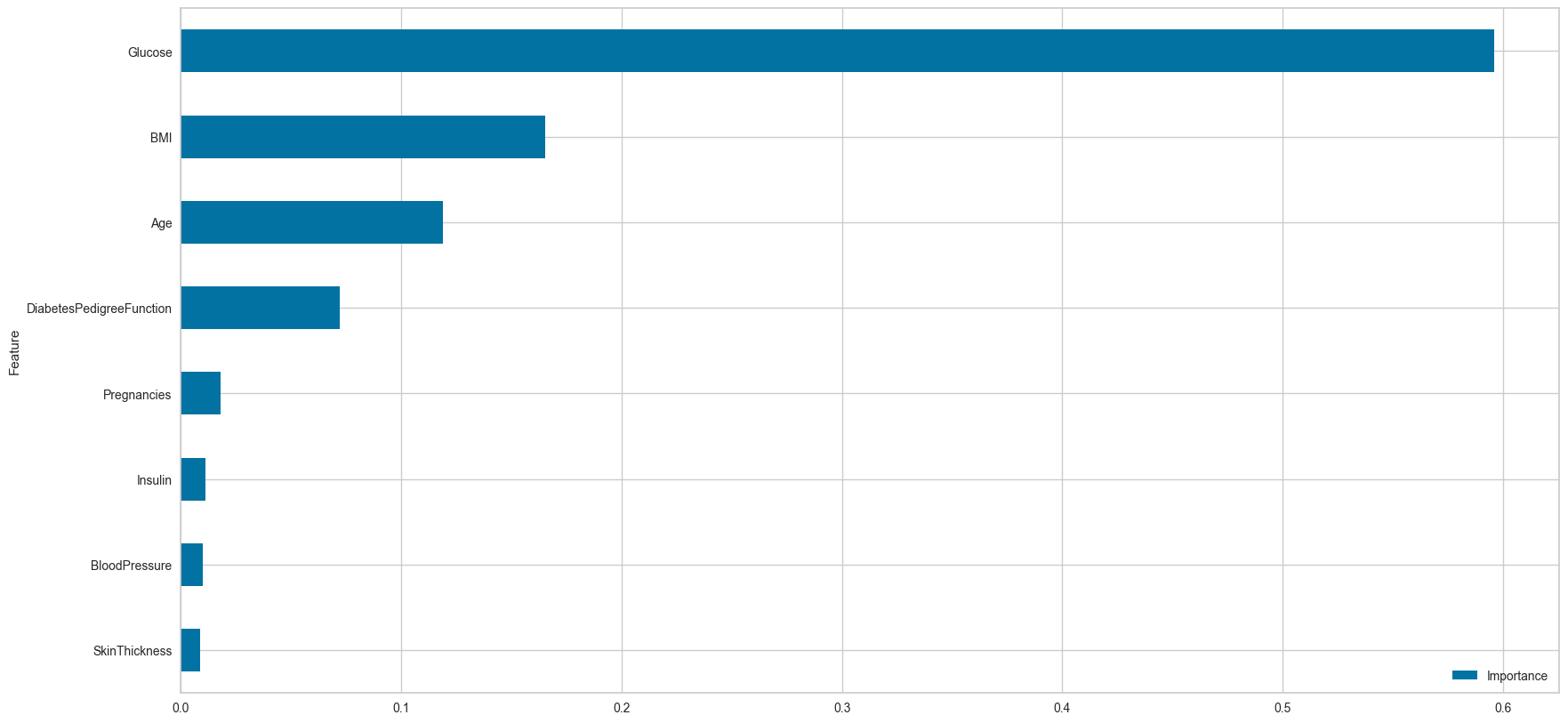
Insulin is not an impact feature for this dataset. Digging deeper on the source of the data, there’s a description that said: “This population has minimal European admixture, and their diabetes appears to be exclusively type 2 diabetes, with no evidence of the autoimmunity characteristic of type 1 diabetes”. Type 2 diabetes patients can still produce insulin, but the body is unable to use it. This information about insulin can be useful later if we want to perform feature selection, in order to boost this model’s performance
Partial Dependency Plot
num_features = X_train_processed.columns.tolist()
num_features['Pregnancies',
'Glucose',
'BloodPressure',
'SkinThickness',
'Insulin',
'BMI',
'DiabetesPedigreeFunction',
'Age']pdp_numerical_only(final_model,X_train_processed,num_features,class_names=['no diabetes','diabetes'],
nrows=4,ncols=2)
# y_class is ignored in binary classification or classical regression settings.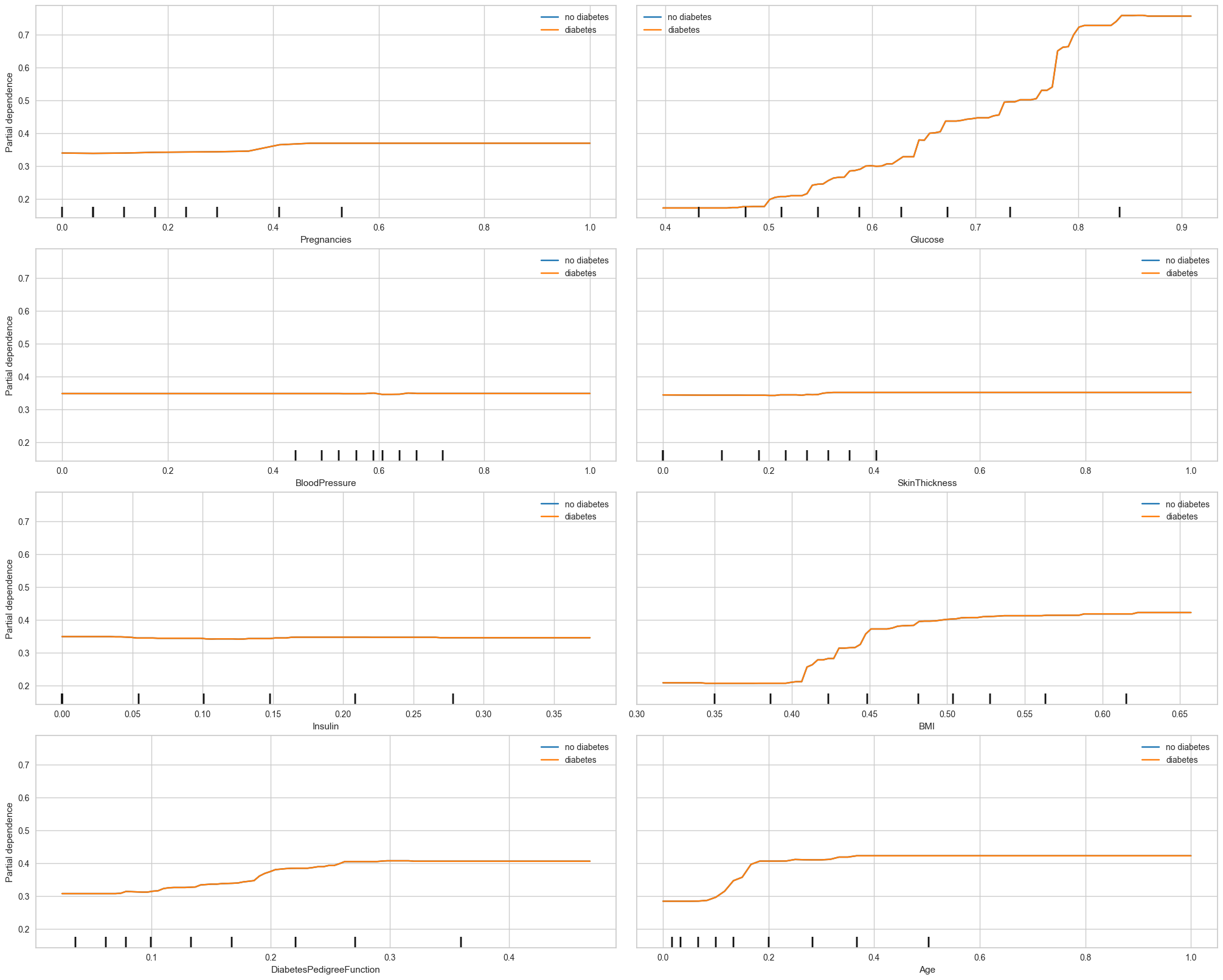
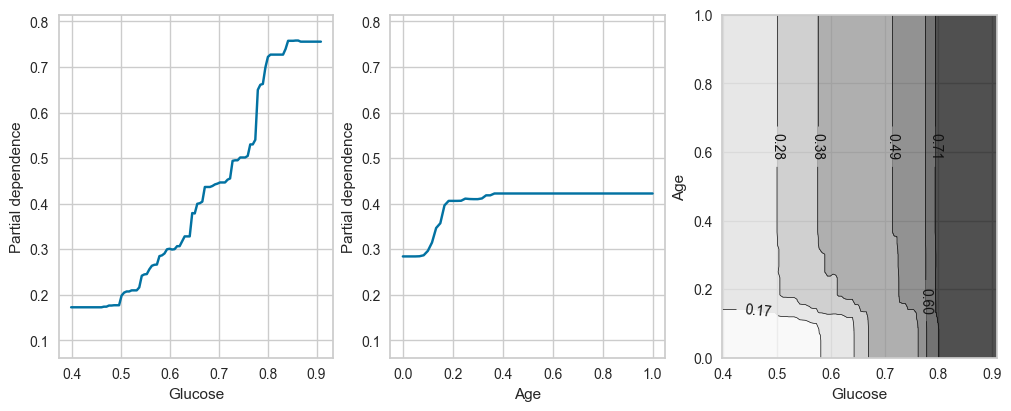
For Partial Dependency Plot, Glucose, BMI and Age show the most changes compared to other features. This matches the feature importances plot earlier.
plot_ice_pair(final_model,X_train_processed,pair_features=['Glucose','Age'],class_idx=1,figsize=(10,4))
# class_idx is ignored in binary classification or classical regression settings.